NumPy is the most popular Python module. It is popular for its N-dimensional array structure and suite of tools that can be used to create, modify, and process them. It also serves as the backbone for data structures provided by other popular modules including Pandas DataFrames, TensorFlow tensors, PyTorch tensors, and many others. Additionally, NumPy is written largely in C, which results in code that runs faster than traditional Python.
What if there were a simple way to find out if your Python code that uses NumPy could be sped up even further? Fortunately, there is!
ndarrays
NumPy, like everything else, stores its data in memory. When a NumPy ndarray is written to memory, its contents are stored in row-major order by default. That is, elements in the same row are adjacent to one another in memory. This order is known as C contiguous, since it's how arrays are stored in memory by default in C.
import numpy as np
x = np.array([[1, 2, 3, 4, 5], [6, 7, 8, 9, 10]])
print(x)
print(x.flags['C_CONTIGUOUS'])
In this case, each element of x is adjacent to its row neighbors in memory. Since memory can be visualized as a flat buffer, it looks like this:
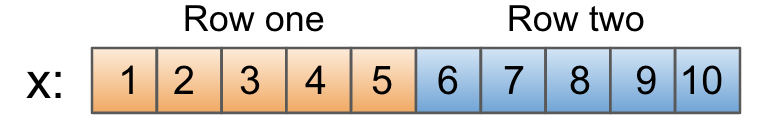
That's straightforward enough. But did you know that you can set the contiguity of a NumPy array yourself? The other common contiguity is known as F contiguous, since it's how arrays are stored in memory by default in Fortran.
import numpy as np
y = np.asfortranarray(x)
print(x.flags['F_CONTIGUOUS'])
In this case, each element of y is adjacent to its column-wise neighbors in memory. That looks like this in a flat buffer:
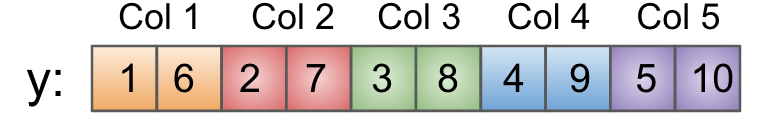
Examples
But what difference does the storage method make in terms of your Python code? It turns out that it can make a significant difference in terms of speed, depending on the dimensions of your data and the operations you want to perform. Code execution time decreases as values in memory get closer together.
Let's explore this with some simple examples.
import numpy as np
import time
row_major_array = np.random.uniform(0, 1, (10000, 2))
col_major_array = np.asfortranarray(np.array(row_major_array, copy = True))
print(row_major_array.flags['C_CONTIGUOUS'])
print(col_major_array.flags['F_CONTIGUOUS'])
start = time.time()
for i in range(1000):
row_major_sum_along_rows = np.sum(row_major_array, axis = 0)
row_major_sum_along_columns = np.sum(row_major_array, axis = 1)
end = time.time()
row_major_elapsed = (end - start) / 1000
start = time.time()
for i in range(1000):
col_major_sum_along_rows = np.sum(col_major_array, axis = 0)
col_major_sum_along_columns = np.sum(col_major_array, axis = 1)
end = time.time()
col_major_elapsed = (end - start) / 1000
print(f"Row major average time: {row_major_elapsed} seconds.")
print(f"Col major average time: {col_major_elapsed} seconds.")
We construct row_major_array and col_major_array, which are each two-dimensional arrays with 2 columns and 10,000 rows of random data between 0 and 1. The contiguity of the arrays are set in accordance with their naming. Then, the column-wise and row-wise sums are computed. We perform each of the two summations 1,000 times and time it, then divide the total time elapsed by 1,000 to see what the average computation time is.
Here, column major order is faster than row major order. The memory ordering of col_major_array is such that the distance between subsequent values needed for the summations is much smaller on average. This difference is significant. Let's try a similar operation on an array with far more rows than columns.
import numpy as np
import time
row_major_array = np.random.uniform(0, 1, (2, 10000))
col_major_array = np.asfortranarray(np.array(row_major_array, copy = True))
print(row_major_array.flags['C_CONTIGUOUS'])
print(col_major_array.flags['F_CONTIGUOUS'])
start = time.time()
for i in range(1000):
row_major_sum_along_rows = np.sum(row_major_array, axis = 0)
row_major_sum_along_columns = np.sum(row_major_array, axis = 1)
end = time.time()
row_major_elapsed = (end - start) / 1000
start = time.time()
for i in range(1000):
col_major_sum_along_rows = np.sum(col_major_array, axis = 0)
col_major_sum_along_columns = np.sum(col_major_array, axis = 1)
end = time.time()
col_major_elapsed = (end - start) / 1000
print(f"Row major average time: {row_major_elapsed} seconds.")
print(f"Col major average time: {col_major_elapsed} seconds.")
This time around, each array has 2 rows and 10,000 columns. The same two summations are performed. Unsurprisingly, the performance difference is approximately equal but opposite to our first example. This time around, the row major order performs better than column major due to the smaller memory distance traveled to fetch required values.
For Python applications that deal with relatively small historical data, these speed differences will not make a major difference in performance. But if you deal with sufficiently large datasets, or high volumes of data coming in real time, these speed differences can have a huge impact. In both of the presented examples, array contiguity is solely responsible for an approximately 10x difference in speed. These are toy problems; the actual performance differences will vary from application to application. NumPy does give you the ability to specify array contiguity for a reason, though!
If there are other ways you speed up your Python code that uses NumPy, we'd love to hear about it. Join our Slack community where we enjoy discussion of topics like this one.

