Deephaven is commonly used to manipulate huge amounts of data — multiple tables, billions of rows, and hundreds of columns. It's built to excel in both versatility of operations and speed of execution.
In this article, we'll explore how Deephaven optimizes memory usage for rapid grouping and ungrouping, and then cover how choosing the right selection method can change the execution time of operations by orders of magnitude. Finally, we'll delve into strategies you can employ to write the most efficient queries possible.
Deephaven saves time automatically where possible, but it's still up to you to write efficient queries. We'll offer techniques — both simple and more complex — to help you maximize the speed of your Deephaven queries.
Built for speed
Data often needs to be organized and sorted before performing any of Deephaven's complex table operations. That means a lot of moving individual pieces of data in and out of various groups.
Whether grouping or ungrouping, moving large amounts of data around has to be a lot of work, and thus take a lot of time. After all, every bit of data being moved has to be copied from one place to another. Right? Not quite.
Deephaven has loads of built-in optimizations that eliminate redundant operations, resulting in massive performance boosts for computationally expensive queries.
For instance, Deephaven often accomplishes grouping and ungrouping via smart indexing operations instead of brute-force copying. Here's an example of Deephaven's speed when grouping and ungrouping 100 columns of 10 million rows:
Time to group: 482 milliseconds.
To avoid some optimizations that happen under the hood and get an accurate time for the ungrouping operation, we'll have to create a new worker for the second half of this test. Then, we'll use that worker to read in and then ungroup the data that we just grouped in the previous code block.
Time to ungroup: 19 milliseconds.
For testing, the code above was run 10 times, each from a fresh Deephaven Community Core worker with 4GB of RAM. Here are the averages from those 10 runs:
- 431 milliseconds to group the data, or 43.1 nanoseconds per row.
- 20 milliseconds to ungroup the data, or 2 nanoseconds per row.
Deephaven was restarted between each run of both code blocks above in order to avoid optimizations that would skew results. In this case, the query engine memoizes the grouping and ungrouping operations results, eliminating redundant calculations. If the Deephaven worker wasn't restarted in between runs, measurements of execution time would be inaccurate, as the engine performs significantly less work when the memoized result is available after the first timed run.
Compute data immediately or as-needed
In the previous example, we used update to create new columns. The update method immediately computes and stores all of the new column values.
The update_view table operation produces identical results to update, but only stores the formula used for the new column(s). Calculations are then performed on-demand when the UI needs to display results or when results are required for a downstream operation. These results are never saved to memory - update_view will have to recalculate the results every time they are needed. This can be a huge time-saver if the results are needed infrequently, but can take much longer downstream than update if the results are needed many times.
We can draw a comparison between update_view and the video game design practice of frustum culling. In a video game, the player can only see a small portion of the game world at any given time. The space within the player's field of view is the frustum, and frustum culling is the practice of selectively rendering only the portion of the game world that the player can see.
Modern video games with massive in-game worlds heavily rely on frustum culling to reduce loading times, and Deephaven queries are no different - they may greatly benefit from deferred calculations.
Let's use the following two examples to compare update and update_view. To ensure that we don't run out of heap space, we will use a table with only 135 million rows. Using new, separate containers, we will perform a summing operation:
- once with grouping using
update - once with grouping using
update_view
The code for this example has been collapsed for readability. Click here to view the code.
Note
The next two code blocks are run separately, on fresh Deephaven workers, to ensure that the results are not skewed by memoization.
Deephaven took 0.22206854820251465 seconds to perform the aggregation on 135 million rows of ints, using `update_view`.
Deephaven took 16.491493701934814 seconds to perform the aggregation on 135 million rows of ints, using `update`.
By repeating the above tests ten times each — again, with a fresh Deephaven worker each time — we get the following average results:
- 0.26 seconds using
update_view- 1.94 nanoseconds per row. - 17.93 seconds using
update- 132.8 nanoseconds per row.
As you can see, the aggregation using update_view is significantly faster than the aggregation using update. This is because update_view only computes the values that are within the view of the UI or are needed by downstream operations. For example, in the code above, update_view doesn't have to perform the calculations right away — so each row completes almost instantly. In contrast, update computes all of the values immediately and stores them in memory.
Let's do one last comparison to see how these two queries affect the performance of downstream operations. We'll re-run the two above code blocks, and add then time two additional operations:
Deephaven took 40.14136099815369 seconds to perform the downstream operations using `update_view`.
Deephaven took 0.019984722137451172 seconds to perform the downstream operations using `update`.
By repeating this test ten times each with the aggregated_update tables from the previous two code blocks, we get the following results:
- 36.20 seconds on the table created using
update_view- 268.16 nanoseconds per row. - 0.016 seconds on the table created using
update- 0.12 nanoseconds per row.
The operation completes almost instantly on the table that we created using update, because the previous calculations have already been done. The operation takes more than two thousand times longer on the table that we created using update_view, because the calculations were deferred and instead have to be performed on-demand every time a downstream operation requires results from the original query.
If we combine the times for both parts of the above test, we see that the overall time to complete the entire operation is 36.46 seconds for the table created using update_view and 17.95 seconds for the table created using update. While the table created using update_view is faster to create, it is significantly slower to use in downstream operations - and the more downstream operations there are, the more pronounced the performance difference between update and update_view will be. Meanwhile, it takes 20 seconds to create the source table using update, but the downstream operations complete almost instantly.
Tip
Choosing the right selection method is a simple way to streamline your queries. See our full guide for choosing a selection method here.
Writing faster queries
Deephaven saves time automatically where possible, but it's still up to you to write efficient queries. Large data sets amplify the effect of small inefficiencies in queries, resulting in significant performance degradation. If you're working with real-time or large data, query efficiency quickly becomes paramount. See our guide Think Like a Deephaven Ninja for tips on writing efficient queries.
Beyond making sure you've chosen the correct selection method for your use case, you may be able to further improve query efficiency by changing the way you represent your logic as a function.
In the examples below, we can see up to a 7x difference in speed depending on how we write our query. We'll break it down into parts, as it's pretty complex.
First, we'll import all the libraries we need.
Click here to view imports.
Next, we'll define a function that groups and aggregates data. This function will be used in all of our examples.
Now, we want to compare several different ways of writing a function for our query. We'll define functions that calculate its first input plus the square root of its second input. The functions are:
custom_func_python, which uses Python's built-in operations.custom_func_numpy, which uses NumPy.custom_func_numba, which uses Numba'sguvectorizedecorator.custom_func_numbanumpy, which uses Numba'sguvectorizedecorator and NumPy.
Note
Deephaven's implementation of guvectorize does not currently use advanced features like chunking, which means it won't be as fast as it could be in this example. Improvements to guvectorize are planned for future releases.
We're almost ready, but we'll need a way to time the above functions as they run. To do that, we define a function that calls each in a table operation, and time the results:
Now, we need to define a few parameters to feed the function we just made:
n_rowsis a list of the number of rows to use in our test.n_groupsis a list of the number of groups to use in our test.n_repeatis the number of times to repeat the test.
We'll also create an empty array to hold our result data:
It's time to put everything together. In the next code block, we will create a loop that runs each of our functions on each of our test cases. Lastly, a table is produced that shows the performance data. We use format_columns to create a heatmap of the results, with the fastest results in green and the slowest in dark gray.
To view the entire above example in one code block, click here.
By repeating the above tests ten times each — again, with a fresh Deephaven worker each time — we get the following average results:
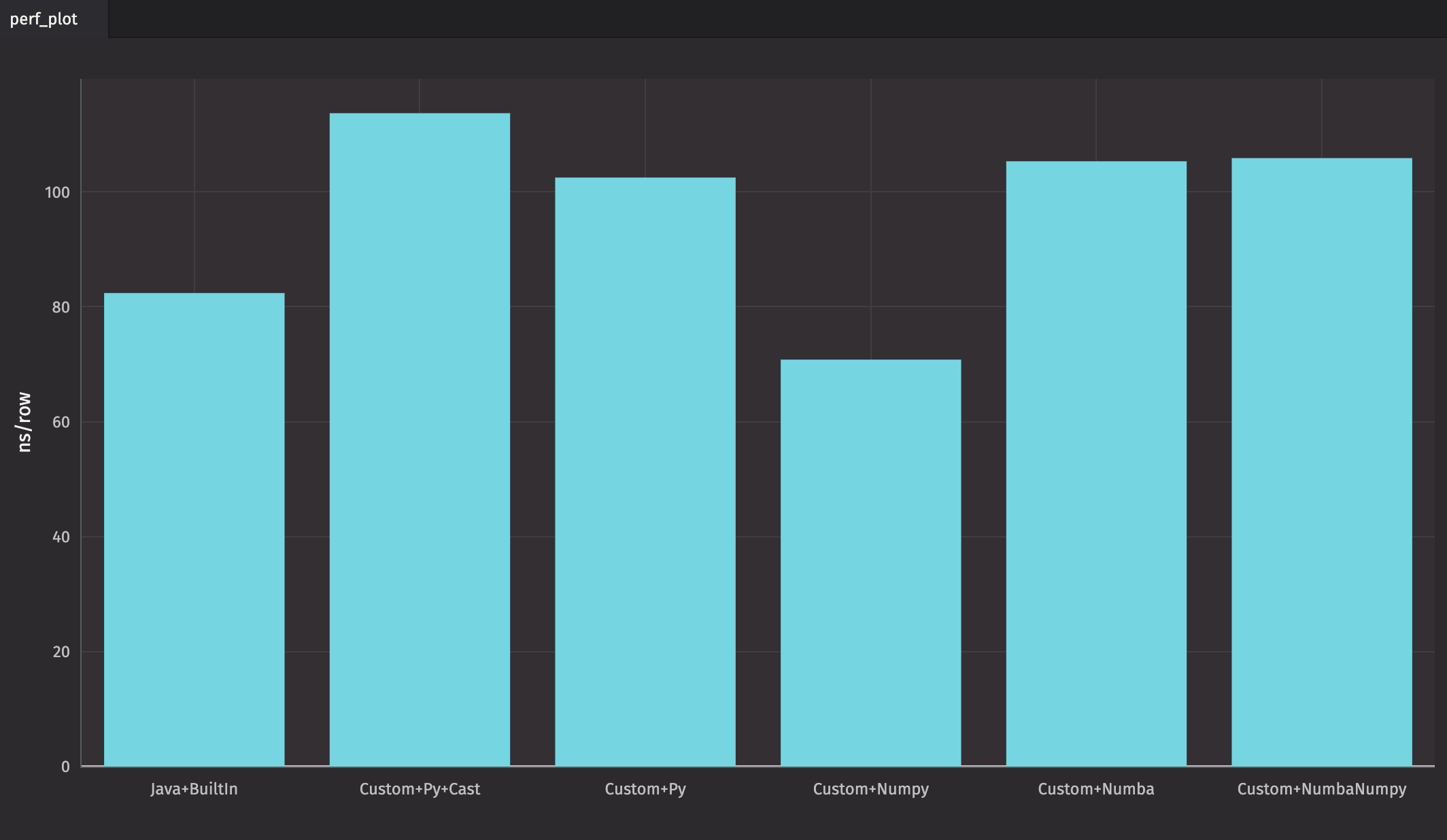
As you can see, "Custom+Numpy" is the fastest in this case, followed closely by the query that uses Java and Deephaven's built-in methods. However, queries that use a combination of Java and built-in methods are almost always the fastest.
Reach out
Our Community documentation has all of the resources you need to become a Deephaven power user. Our Slack community continues to grow, and we'd love to have you join us! If you have any questions, comments, or suggestions, please reach out to us there.

