Use SciPy
This guide will show you how to use SciPy in your Python queries in Deephaven.
Refer to our guide How to install Python packages for instructions on installing this package in Deephaven.
SciPy is an open-source scientific computing library for Python. It contains a large assortment of sub-packages with methods that can perform tasks in the following areas:
- Optimization
- Linear algebra
- Calculus
- Interpolation
- Transforms
- Signal processing
- Differential equations
- Multi-dimensional array processing
The above list is not comprehensive; SciPy's many sub-packages cover an even broader range of topics (see the Appendix at the bottom of this guide for a brief description of each). SciPy was built to operate on NumPy arrays, which we'll be using extensively in this guide. Check out How to use NumPy to learn more about how they work.
Examples
The examples below demonstrate using SciPy in a traditional Python setting, then extend them to use Deephaven. SciPy is a natural pair for deephaven.learn, so some of the corresponding Python queries with Deephaven use it. For more information on the learn submodule, check out How to use deephaven.learn.
Describe data
In this example, we'll use scipy.stats.describe to give basic insight into some data. The data we'll be describing is an ndarray with integer values from 0 to 99.
from scipy.stats import describe
import numpy as np
source = np.arange(100)
result = describe(source)
print(result)
To get these insights on data in a Deephaven table, we'll convert it to a NumPy array by using pandas.to_pandas and values. We can then use the method just as before.
from deephaven import empty_table, pandas
from scipy.stats import describe
import pandas as pd
import numpy as np
source = empty_table(100).update(formulas=["X = i"])
result = describe(np.squeeze(pandas.to_pandas(source).values))
print(result)
![]()
Compute the Airy function
In this example, we'll use scipy.special.airy to compute the Airy function of some values. In this case, we'll calculate the Airy function on values ranging from -20 to 0 in increments of 0.2.
from scipy.special import airy
import numpy as np
x = np.linspace(-20, 0, 101)
ai, aip, bi, bip = airy(x)
Let's extend this code to use Deephaven tables!
from deephaven import empty_table
from scipy.special import airy
import numpy as np
source = empty_table(101).update(formulas=["x = -20 + 20 * i / 100"])
def compute_airy(x):
ai, aip, bi, bip = airy(x)
return ai, aip, bi, bip
result = source.update(formulas=["y = compute_airy(x)"])
Filter a noisy signal
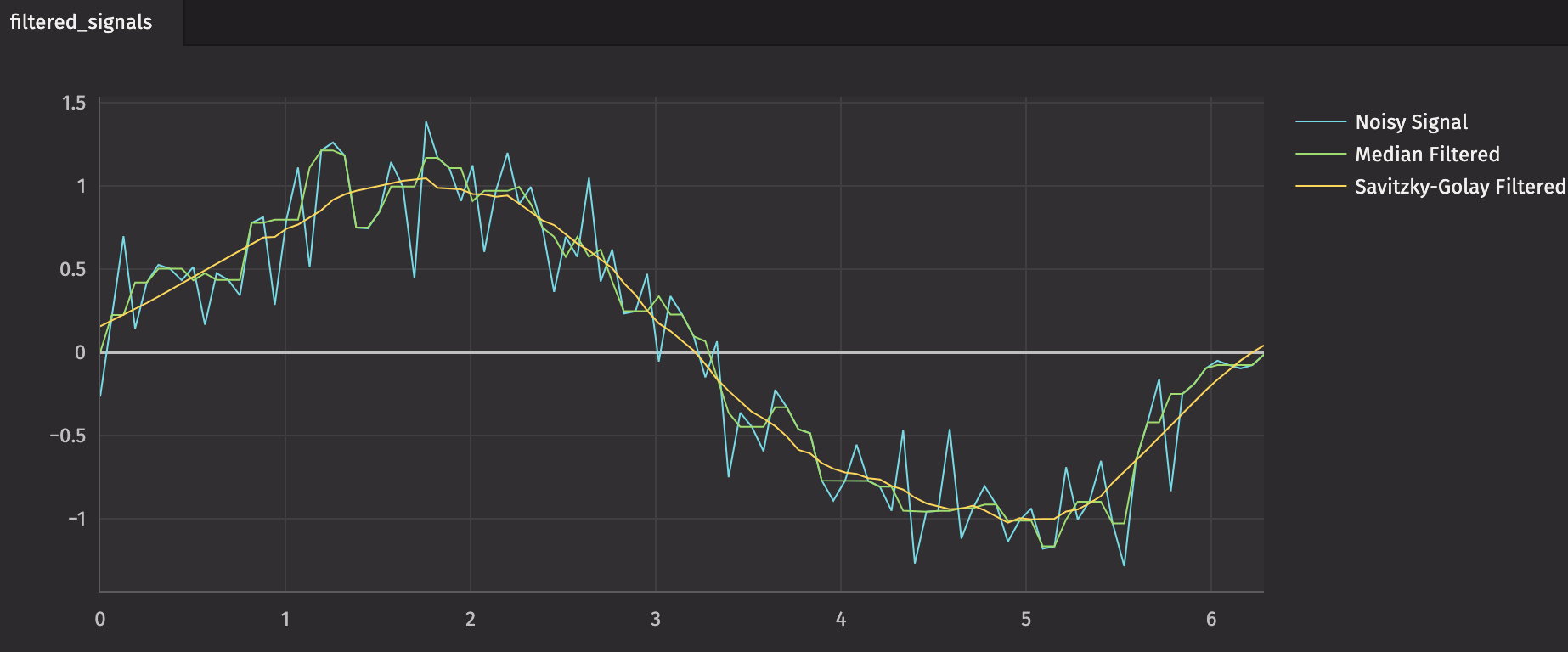
In this example, we'll generate a Sine wave and add noise. Then, we'll apply filters to reduce the noise.
from scipy.signal import medfilt, savgol_filter
import numpy as np
x = np.linspace(0, 2 * np.pi, 101)
noisy_sine_wave = np.sin(x) + np.random.normal(0, 0.25, 101)
median_filtered = medfilt(noisy_sine_wave)
savgol_filtered = savgol_filter(noisy_sine_wave, 27, 3)
The above example can be extended to use deephaven.learn in a couple of ways.
from deephaven.plot.figure import Figure
from deephaven.learn import gather
from deephaven import empty_table
from deephaven import learn
from scipy.signal import medfilt, savgol_filter
import numpy as np
def create_noisy_sine_wave(x) -> np.double:
return np.sin(x) + np.random.normal(0, 0.25)
source = empty_table(101).update(
formulas=[
"X = i * (2 * Math.PI) / 100",
"Noisy_Sine_Wave = create_noisy_sine_wave(X)",
]
)
def apply_median_filter(signal):
return medfilt(signal)
def apply_savgol_filter(signal):
return savgol_filter(signal, 27, 3)
def table_to_numpy(rows, cols):
return np.squeeze(gather.table_to_numpy_2d(rows, cols, np_type=np.double))
def numpy_to_table(data, idx):
return data[idx]
median_filtered = learn.learn(
table=source,
model_func=apply_median_filter,
inputs=[learn.Input("Noisy_Sine_Wave", table_to_numpy)],
outputs=[learn.Output("Median_Filtered", numpy_to_table, "double")],
batch_size=source.size,
)
savitzky_golay_filtered = learn.learn(
table=source,
model_func=apply_savgol_filter,
inputs=[learn.Input("Noisy_Sine_Wave", table_to_numpy)],
outputs=[learn.Output("Savitzky_Golay_Filtered", numpy_to_table, "double")],
batch_size=source.size,
)
plt = (
Figure()
.plot_xy(series_name="Noisy Signal", t=source, x="X", y="Noisy_Sine_Wave")
.plot_xy(
series_name="Median Filtered", t=median_filtered, x="X", y="Median_Filtered"
)
.plot_xy(
series_name="Savitzky-Golay Filtered",
t=savitzky_golay_filtered,
x="X",
y="Savitzky_Golay_Filtered",
)
.show()
)
Compute the nearest neighbor of every point in a set
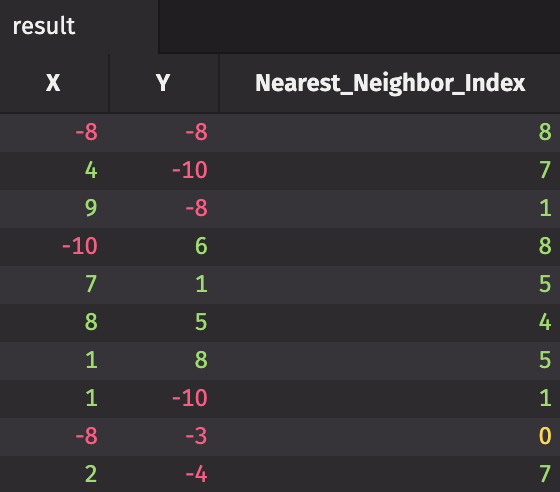
In this example, we'll use a K-d tree to compute the nearest neighbor of every point in a set. SciPy's implementation is called scipy.spatial.KDTree. We construct a two-dimensional data set with a few points, and we'll find the index of each point's nearest neighbor in the set.
from scipy.spatial import KDTree as kdtree
import numpy as np
x = np.random.randint(-10, 10, 10)
y = np.random.randint(-10, 10, 10)
data = np.vstack((x, y)).T
print(data)
tree = kdtree(data)
distances, indices = tree.query(data, k=2)
nearest_neighbor_indices = indices[:, 1]
print(nearest_neighbor_indices)
Computing the nearest-neighbor distances and indices can also be done using deephaven.learn.
from deephaven.pandas import to_table
from deephaven.learn import gather
from deephaven import learn
from scipy.spatial import KDTree as kdtree
import pandas as pd
import numpy as np
indices = np.arange(10)
x = np.random.randint(-10, 10, 10)
y = np.random.randint(-10, 10, 10)
source = to_table(pd.DataFrame({"Index": indices, "X": x, "Y": y}))
def calculate_nearest_neighbor_indices(data):
tree = kdtree(data)
distances, indices = tree.query(data, k=2)
return indices[:, 1]
def table_to_numpy(rows, cols):
return gather.table_to_numpy_2d(rows, cols, np_type=np.int_)
def numpy_to_table(data, idx):
return data[idx]
result = learn.learn(
table=source,
model_func=calculate_nearest_neighbor_indices,
inputs=[learn.Input(["X", "Y"], table_to_numpy)],
outputs=[learn.Output("Nearest_Neighbor_Index", numpy_to_table, "int")],
batch_size=source.size(),
)
Compute distances of various metrics between two vectors
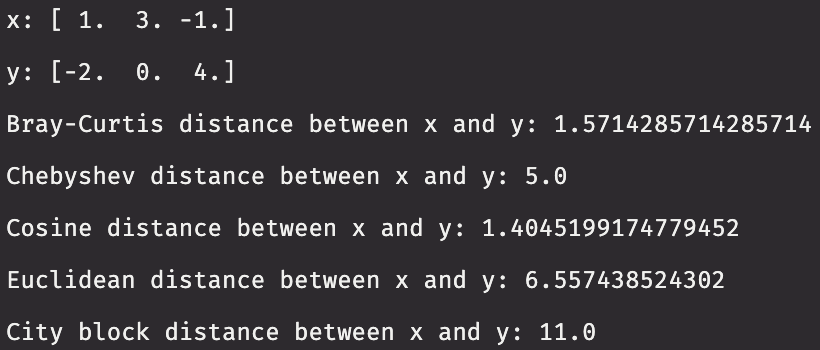
In this example, we create two three-dimensional vectors. We then compute the distance between the two vectors using metrics supported by scipy.spatial.distance.
from scipy.spatial import distance
import numpy as np
x = np.array([1, 3, -1])
y = np.array([-2, 0, 4])
def print_distances(x, y):
print("x: " + str(x))
print("y: " + str(y))
print("Bray-Curtis distance between x and y: " + str(distance.braycurtis(x, y)))
print("Chebyshev distance between x and y: " + str(distance.chebyshev(x, y)))
print("Cosine distance between x and y: " + str(distance.cosine(x, y)))
print("Euclidean distance between x and y: " + str(distance.euclidean(x, y)))
print("City block distance between x and y: " + str(distance.cityblock(x, y)))
print_distances(x, y)
We can once again use deephaven.learn to calculate these distances.
from deephaven import new_table, learn
from deephaven.column import int_col
from deephaven.learn import gather
from scipy.spatial import distance
import numpy as np
source = new_table([int_col("X", [1, 3, -1]), int_col("Y", [-2, 0, 4])])
def table_to_numpy(rows, cols):
return np.squeeze(gather.table_to_numpy_2d(rows, cols, np_type=np.intc))
def numpy_to_table(data, idx):
return data[idx]
def print_distances(data):
x = data[:, 0]
y = data[:, 1]
print("x: " + str(x))
print("y: " + str(y))
print("Bray-Curtis distance between x and y: " + str(distance.braycurtis(x, y)))
print("Chebyshev distance between x and y: " + str(distance.chebyshev(x, y)))
print("Cosine distance between x and y: " + str(distance.cosine(x, y)))
print("Euclidean distance between x and y: " + str(distance.euclidean(x, y)))
print("City block distance between x and y: " + str(distance.cityblock(x, y)))
learn.learn(
table=source,
model_func=print_distances,
inputs=[learn.Input(["X", "Y"], table_to_numpy)],
outputs=None,
batch_size=source.size(),
)
Appendix: Sub-packages of SciPy
Here is a comprehensive list of the sub-packages available in SciPy, and a brief description of what each one does:
| Sub-package name | Description |
|---|---|
cluster | Clustering routines |
constants | A collection of constants and conversion utilities |
fft | Fourier transform methods |
fftpack | Deprecated legacy Fourier transform methods |
ingegrate | Numerical integration algorithms |
interpolate | Tools for data interpolation |
io | Data I/O |
linalg | Linear algebra routines |
misc | Extra miscellaneous utilities |
ndimage | Image processing library |
odr | Orthogonal distance regression library |
optimize | Algorithms for optimizing code |
signal | Signal processing routines |
sparse | Tools for sparse matrix creation, handling, and procesing |
spatial | Spatial structure processing algorithms |
special | Special functions |
stats | Statistical function library |
weave | Deprecated legacy C/C++ code writing tool |
Each is described below. We skip fftpack and weave because they are deprecated.## Appendix
scipy.cluster
scipy.cluster is split into two further sub-modules:
scipy.cluster.vqsupports vector quantization and K-means algorithms.scipy.cluster.heirarchyprovides hierarchical and agglomerative clustering.
scipy.constants
scipy.constants provides an extensive number of mathematical and physical constants including (but not limited to) pi, the golden ratio, the Planck constant, subatomic particle masses, SI prefixes, binary prefixes, and many others. The full list of constants is far, far too long to include here.
scipy.fft
scipy.fft contains methods that apply the forward and reverse discrete Fourier, Sine, Cosine, and Hankel transformations to input data in N dimensions.
scipy.integrate
scipy.integrate contains a slew of methods to calculate integrals and solutions for differential equations.
scipy.interpolate
scipy.interpolate is a sub-package for objects used in data interpolation. It provides univariate and multivariate interpolation techniques, and spline interpolators.
scipy.io
scipy.io provides utilities to read and write data to and from various file formats including MATLAB, Fortran, WAV, and others.
scipy.linalg
scipy.linalg is a library of linear algebra functions. You can use this sub-module to solve eigenvalue problems, perform matrix decompositions, solve matrix equations, and construct special matrices among other things. Matrices constructed using this sub-package are of type numpy.ndarray.
This sub-module contains two further sub-packages for low-level routines: scipy.linalg.blas and scipy.linalg.lapack. These two sub-packages provide low-level functions from the Basic Linear Algebra Subprograms (BLAS) and Linear Algebra Package (LAPACK) libraries. Cython implementations of these low-level libraries exist as well.
Additionally, it provides routines for interpolative matrix decomposition via scipy.linalg.interpolative.
scipy.misc
The miscellaneous routines sub-module, scipy.misc, is the home of some example data sets and two special derivative calculators.
scipy.ndimage
scipy.ndimage contains a library of methods to perform image processing. Methods include filters, interpolation routines, measurements, and morphology routines.
scipy.odr
scipy.odr contains routines to perform orthogonal distance regression (ODR) on input data. ODR extends least squares fitting, which generally results in more accuracy in fitted curves.
scipy.optimize
The scipy.optimize sub-module provides univariate and multivariate optimization routines. These optimization routines will minimize or maximize objective functions, and can handle constraints on said functions. Solvers for both linear and nonlinear problems is supported.
There are underlying C functions for four different root finders that can be accessed via Cython.
scipy.signal
scipy.signal is SciPy's signal processing sub-module. It supports convolutions, B-splines, filters, filter designing routines, continuous time linear systems, discrete time linear systems, waveform generators, window functions, wavelet generators, maxima/minima finers, and spectral analyzers.
scipy.sparse
scipy.sparse contains functions for building and identifying sparse matrices in various formats. Additionally, it contains sparse matrix classes for each individual sparse matrix format.
This sub-module contains two sub-packages: scipy.sparse.csgraph and scipy.sparse.linalg. The first contains a library of compressed sparse graph routines, and the second contains a library of sparse linear algebra routines.
scipy.spatial
scipy.spatial contains spatial algorithms and data structures. Features include nearest-neighbor query routines, triangulation, convex hulls, Voronoi diagrams, and plotting helpers.
One sub-package of scipy.spatial is scipy.spatial.distance, which provides utilities for computing distance matrices, cheing their validity, and distance functions in numeric and boolean vectors.
There is another sub-pacakage within scipy.spatial called scipy.spatial.transform, which provides functions for spatial transformations including rotations, spherical linear interpolations of rotations, and rotation splines.
scipy.special
SciPy special functions are defined in scipy.special, which are extended to Cython via scipy.special.cython_special. Special functions include (but are not limited to) Airy, Bessel, Struve, Legendre, and statistical distributions.
scipy.stats
The sub-module, scipy.stats, contains probability distributions, summary statistics, frequency statistics, and functions for calculating correlations between distributions among other things. Methods exist for continuous, discrete, and multivariate distributions. There are are also some summary methods that can be used to describe data with little prior knowledge.
There are two sub-packages within scipy.stats: scipy.stats.mstats and scipy.stats.qmc. The former is used for calculating statistics on masked arrays, and the latter is used for describing quasi-Monte Carlo capabilities.