Use SciKit-Learn in Deephaven
This guide will show you how to use SciKit-Learn in Deephaven queries.
SciKit-Learn is an open-source machine learning library for Python. It features a variety of methods for classification, clustering, regression, and deep learning.
SciKit-Learn does not come stock with Deephaven's base Docker image. To use it within Deephaven, you can install it yourself or choose one of a Deephaven Docker deployments with support built-in. The following options will work:
- Without example data:
- With example data:
Two examples are given below. Both classify observations in the Iris dataset, which can be found in Deephaven's Examples repository. The first example uses SciKit-Learn, whereas the second integrates Deephaven tables to perform predictions on live data
Note
In this guide, we read data from Deephaven's examples repository. You can also load files that are in a mounted directory at the base of the Docker container. See Docker data volumes to learn more about the relation between locations in the container and the local file system.
The Iris flower dataset is a popular dataset commonly used in introductory machine learning applications. Created by R.A. Fisher in his 1936 paper, The use of multiple measurements in taxonomic problems, it contains 150 measurements of three types of Iris flower subspecies. The following values are measured in centimeters for Iris-setosa, Iris-versicolor, and Iris-virginica flowers:
- Petal length
- Petal width
- Sepal length
- Sepal width
This is a classification problem suitable for a classifier algorithm. We'll use the K-Nearest Neighbors classifier.
Classify the Iris dataset
This first example shows how to use SciKit-Learn to classify Iris flowers from measurements.
Let's first import all the packages we'll need.
from sklearn.neighbors import KNeighborsClassifier as knn
import pandas as pd
import numpy as np
Next, we'll import the data, quantize the targets, and split the data into training and testing sets.
# Read and quantize the dataset
iris = pd.read_csv(
"https://media.githubusercontent.com/media/deephaven/examples/main/Iris/csv/iris.csv"
)
iris_mappings = {"Iris-setosa": 0, "Iris-virginica": 1, "Iris-versicolor": 2}
iris["Class"] = iris["Class"].apply(lambda x: iris_mappings[x])
# Split the DataFrame into training and testing sets
iris_shuffled = iris.sample(frac=1)
train_size = int(0.75 * len(iris_shuffled))
train_set = iris_shuffled[:train_size]
test_set = iris_shuffled[train_size:]
# Separate our data into features and targets (X and Y)
X_train = train_set.drop("Class", axis=1).values
Y_train = train_set["Class"].values
X_test = test_set.drop("Class", axis=1).values
Y_test = test_set["Class"].values
Next, we'll construct a K-Nearest-Neighbors classifier with 3 as the number of neighbors. We fit it to the training set.
neigh = knn(n_neighbors=3)
neigh.fit(X_train, Y_train)
Lastly, we'll predict the remaining data points using this classifier.
test_size = len(X_test)
n_correct = 0
for i in range(test_size):
prediction = neigh.predict([X_test[i]])[0]
if prediction == Y_test[i]:
n_correct += 1
accuracy = n_correct / test_size
print(str(accuracy * 100) + "% correct")

Around 95% correct for such a simple solution. Pretty neat!
This example follows a basic formula for using a classifier to solve a classification problem:
- Import, quantize, update, and store data from an external source.
- Define the classification model.
- Fit the classification model to training data.
- Test the model.
- Assess its accuracy.
In this case, a simple classifier is suitable for a simple dataset.
Classify the Iris dataset with Deephaven tables
So we just classified the Iris dataset using K-Nearest Neighbors. That's kind of cool, but wouldn't it be cooler to do it on a live feed of incoming data? Let's do that with Deephaven!
We extend our previous example to train our model on a Deephaven table. This requires some additional code.
First, we import the packages we need.
# Deephaven imports
from deephaven import DynamicTableWriter
import deephaven.dtypes as dht
from deephaven.learn import gather
from deephaven import read_csv
from deephaven import learn
# Machine learning imports
from sklearn.neighbors import KNeighborsClassifier as knn
# Additional required imports
import numpy as np, random, threading, time
Then, we import and quantize our data as before. This time, we'll import it into a Deephaven table.
iris_raw = read_csv(
"https://media.githubusercontent.com/media/deephaven/examples/main/Iris/csv/iris.csv"
)
classes = {}
num_classes = 0
def get_class_number(c) -> np.intc:
global classes, num_classes
if c not in classes:
classes[c] = num_classes
num_classes += 1
return classes[c]
iris = iris_raw.update(formulas=["Class = get_class_number(Class)"])
This time, we'll create functions to train the classifier and use it.
neigh = 0
# Construct and classify the Iris dataset
def fit_knn(X_train, Y_train):
global neigh
neigh = knn(n_neighbors=3)
neigh.fit(X_train, Y_train)
# Use the K-Nearest neighbors classifier on the Iris dataset
def use_knn(features):
if features.ndim == 1:
features = np.expand_dims(features, 0)
predictions = np.zeros(len(features))
for i in range(0, len(features)):
predictions[i] = neigh.predict([features[i]]).squeeze()
return predictions
Now we need some extra functions. The first two gather data from columns in Deephaven tables into NumPy arrays of doubles and integers, respectively, while the last extracts a value from the predictions. The extracted values will be used to create a new column in the output table.
# A function to gather data from columns into a NumPy array of doubles
def table_to_array_double(rows, cols):
return gather.table_to_numpy_2d(rows, cols, np_type=np.double)
# A function to gather data from columns into a NumPy array of integers
def table_to_array_int(rows, cols):
return np.squeeze(gather.table_to_numpy_2d(rows, cols, np_type=np.intc))
# A function to extract a list element and cast to an integer
def get_predicted_class(data, idx):
return int(data[idx])
Now, we can classify the Iris subspecies from these measurements. This time, we'll do it in a Deephaven table using the learn function. The first time we call learn, we fit the to training data. Then, we use the fitted model to classify the the Iris subspecies in the Class column.
# Use the learn function to fit our KNN classifier
learn.learn(
table=iris,
model_func=fit_knn,
inputs=[
learn.Input(
["SepalLengthCM", "SepalWidthCM", "PetalLengthCM", "PetalWidthCM"],
table_to_array_double,
),
learn.Input("Class", table_to_array_int),
],
outputs=None,
batch_size=150,
)
# Use the learn function to create a new table that contains classified values
iris_knn_classified = learn.learn(
table=iris,
model_func=use_knn,
inputs=[
learn.Input(
["SepalLengthCM", "SepalWidthCM", "PetalLengthCM", "PetalWidthCM"],
table_to_array_double,
)
],
outputs=[learn.Output("ClassifiedClass", get_predicted_class, "int")],
batch_size=150,
)
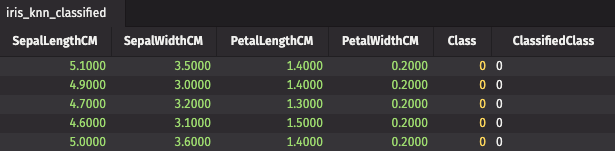
We've done the same thing as before: we created static classifications on static data. Only this time, we used a Deephaven table. That's not that exciting. We really want to perform the classification stage on a live data feed. To demonstrate this, we'll create some fake Iris measurements.
We can create an in-memory real-time table using DynamicTableWriter. To create semi-realistic measurements, we'll use some known quantities from the Iris dataset:
| Column | Minimum (CM) | Maximum (CM) |
|---|---|---|
PetalLengthCM | 1.0 | 6.9 |
PetalWidthCM | 0.1 | 2.5 |
SepalLengthCM | 4.3 | 7.9 |
SepalWidthCM | 2.0 | 4.4 |
These quantities will be fed to our table writer and faux measurements will be written to the table once per second for one minute. We'll apply our model on those measurements as they arrive and predict which Iris subspecies they belong to.
First, we create a live table and write data to it.
# Create the table writer
table_writer = DynamicTableWriter(
{
"SepalLengthCM": dht.double,
"SepalWidthCM": dht.double,
"PetalLengthCM": dht.double,
"PetalWidthCM": dht.double,
}
)
# Get the live, ticking table
live_iris = table_writer.table
# This function creates faux Iris measurements once per second for a minute
def write_to_iris():
for i in range(60):
petal_length = random.randint(10, 69) / 10
petal_width = random.randint(1, 25) / 10
sepal_length = random.randint(43, 79) / 10
sepal_width = random.randint(20, 44) / 10
table_writer.write_row(sepal_length, sepal_width, petal_length, petal_width)
time.sleep(1)
# Use a thread to write data to the table
thread = threading.Thread(target=write_to_iris)
thread.start()
Now we use learn on the ticking table. All it takes to change from static to live data is to change the table!
# Use the learn function to create a new table with live predictions
iris_classified_live = learn.learn(
table=live_iris,
model_func=use_knn,
inputs=[
learn.Input(
["SepalLengthCM", "SepalWidthCM", "PetalLengthCM", "PetalWidthCM"],
table_to_array_double,
)
],
outputs=[learn.Output("LikelyClass", get_predicted_class, "int")],
batch_size=150,
)
All of the code to create and use our trained model on the live, ticking table is below.
# Create the table writer
table_writer = DynamicTableWriter(
{
"SepalLengthCM": dht.double,
"SepalWidthCM": dht.double,
"PetalLengthCM": dht.double,
"PetalWidthCM": dht.double,
}
)
# Get the live, ticking table
live_iris = table_writer.table
# This function creates faux Iris measurements once per second for a minute
def write_to_iris():
for i in range(60):
petal_length = random.randint(10, 69) / 10
petal_width = random.randint(1, 25) / 10
sepal_length = random.randint(43, 79) / 10
sepal_width = random.randint(20, 44) / 10
table_writer.write_row(sepal_length, sepal_width, petal_length, petal_width)
time.sleep(1)
# Use a thread to write data to the table
thread = threading.Thread(target=write_to_iris)
thread.start()
# Use the learn function to create a new table with live classifications
iris_classified_live = learn.learn(
table=live_iris,
model_func=use_knn,
inputs=[
learn.Input(
["SepalLengthCM", "SepalWidthCM", "PetalLengthCM", "PetalWidthCM"],
table_to_array_double,
)
],
outputs=[learn.Output("LikelyClass", get_predicted_class, "int")],
batch_size=150,
)